Epigenetic Moonlighting: Catalytic-Independent Functions
Epigenetic Moonlighting: Catalytic-Independent Functions in Development and Disease Pathogenesis
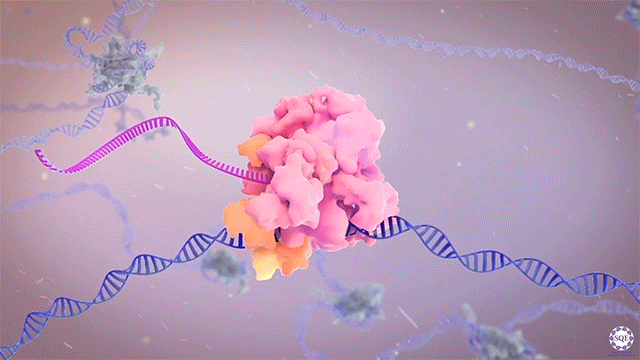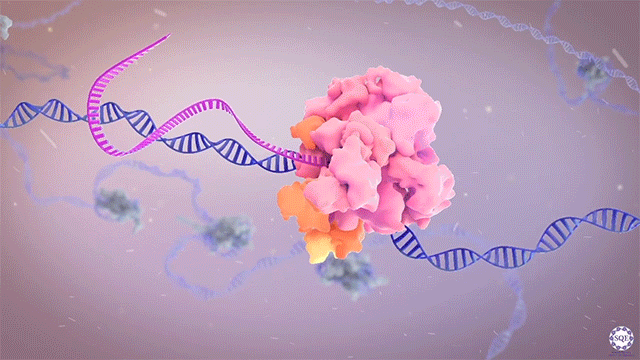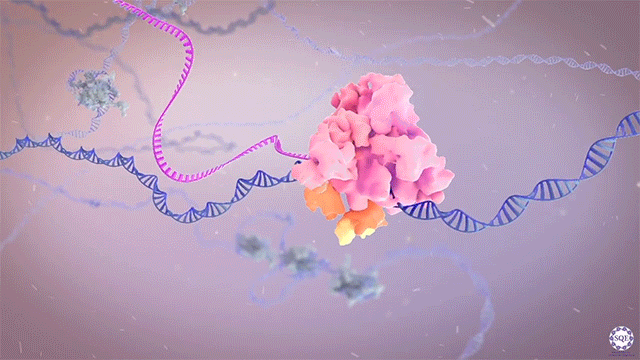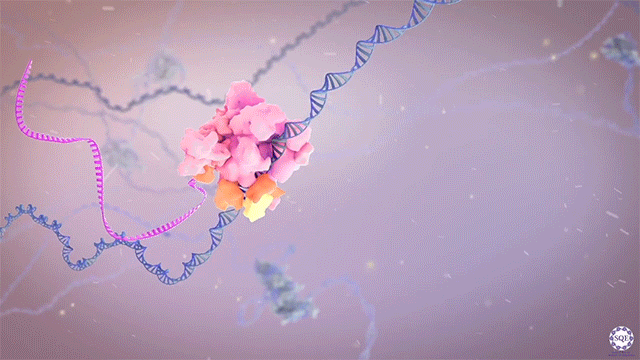
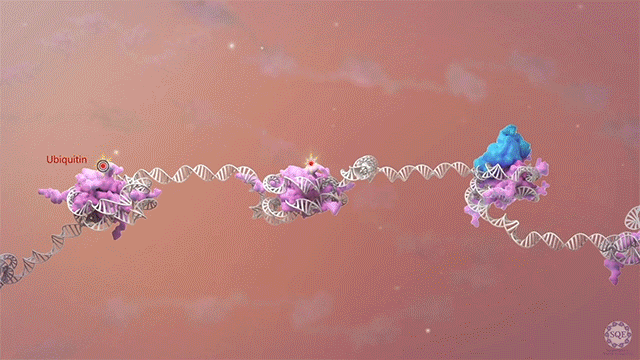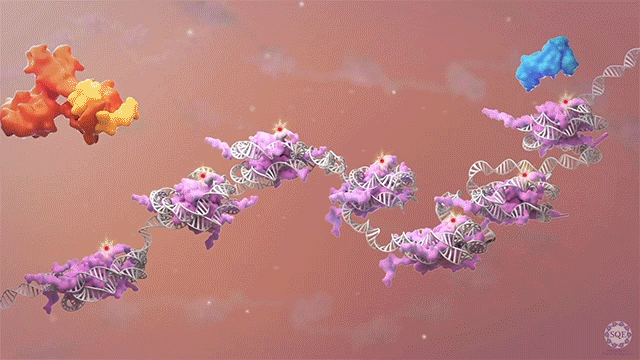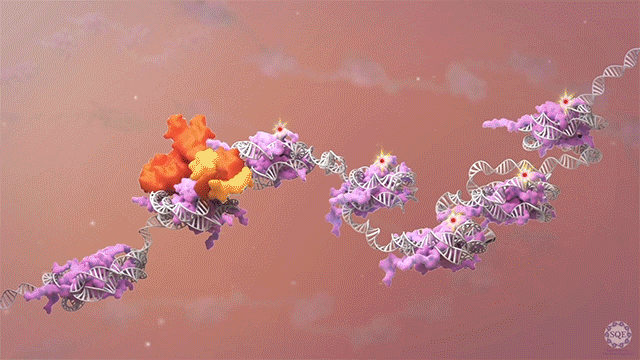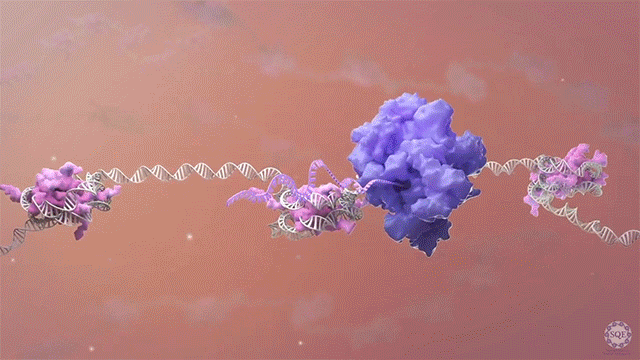
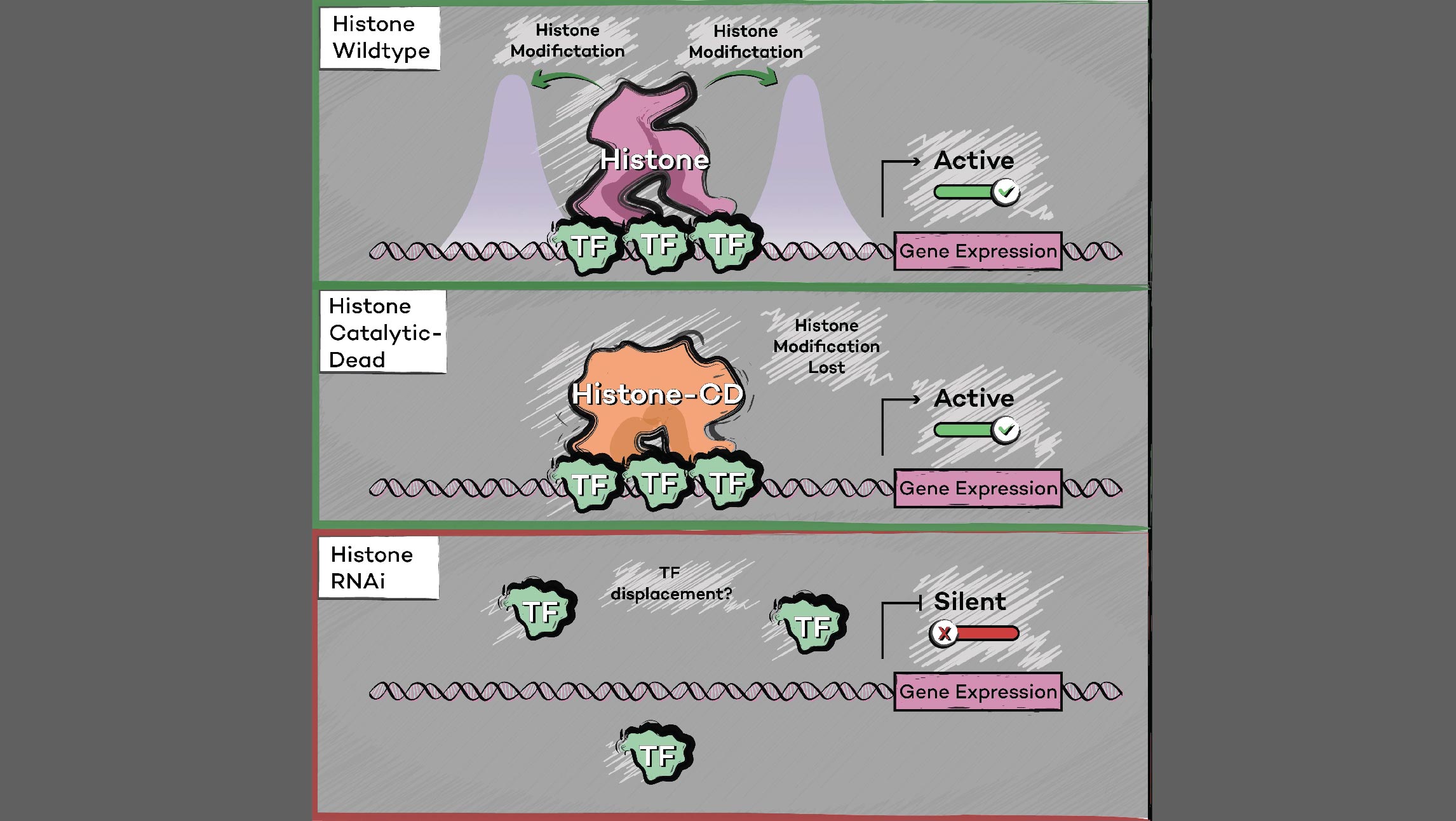
Histone methyltransferases and other chromatin modifiers are often referred to as “writers” of readable and editable marks that constitute a “histone code.” While this prospect is appealing in its simplicity, chromatin function as a whole is enormously complex, and the true role of histone modifications in transcriptional regulation is as yet unclear. Moreover, for a growing list of histone modifiers, we and others have shown that the modifiers can function in the absence of their catalytic activities. These findings indicate critical non-catalytic “moonlighting” functions for certain histone modifying enzymes, and they also prompt a re-evaluation of corresponding histone modifications in the transcriptional regulation of gene expression.
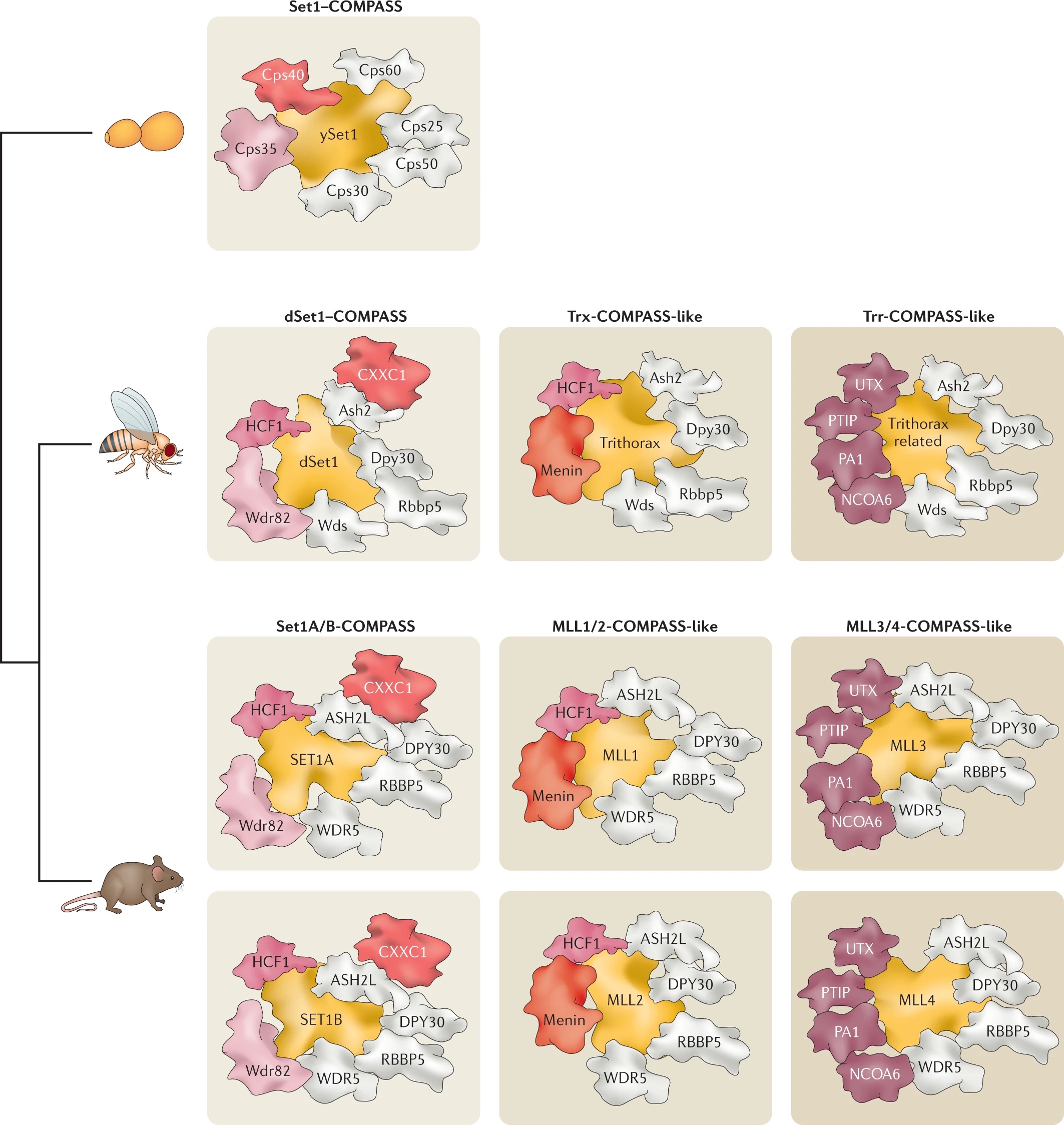
Our laboratory has demonstrated the dispensability of catalytic activity and identified critical non-catalytic functions for several histone modifying enzymes, including COMPASS family members and other methyltransferases. When we introduced inactivating mutations to the catalytic sites of Trr in Drosophila and its mammalian counterparts MLL3/4 in murine embryonic stem cells (mESCs), we found that the resultant loss of H3K4 monomethylation at enhancers did not affect viable development. Moreover, when we created a catalytically hyperactive mutant that increased H3K4 di- and tri-methylation at enhancers, flies with this alternative enhancer histone modification were no less viable.[Rickels 2017] Because the C and N terminal halves of mammalian MLL3/MLL4 are homologous to Drosophila Lpt and Trr, respectively, we were then able to exploit the Lpt/Trr gene split in Drosophila to screen for essential non-catalytic functions of MLL3/4. 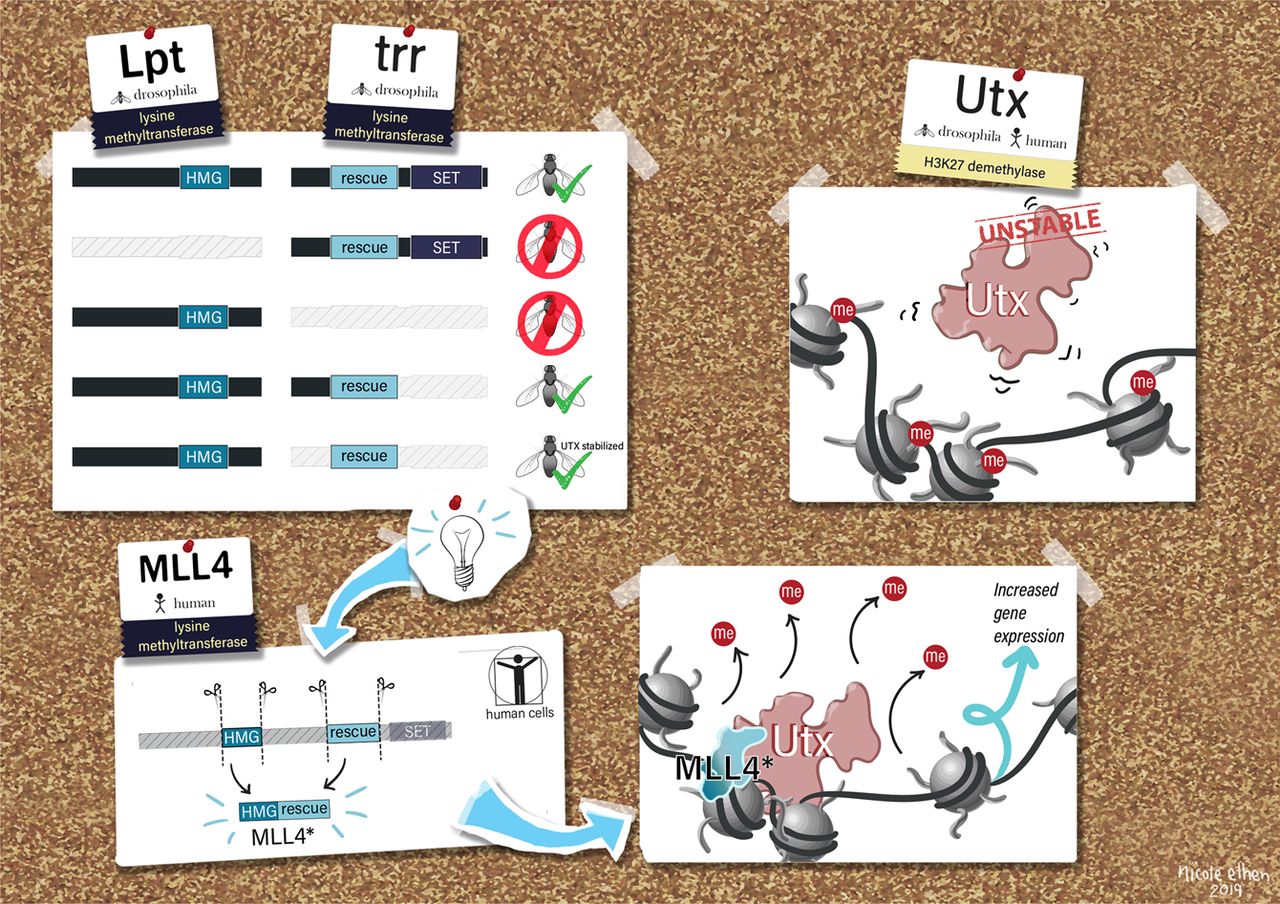
Our screen identified a minimal domain that was sufficient to rescue Trr-null lethality, and we demonstrated that it does so by binding to and stabilizing the H3K27 demethylase and Trr/COMPASS complex member Utx. In mammalian MLL3/4 KO cells, the fusion of an HMG-box (present in C-terminal MLL3/4 and Drosophila homolog Lpt) to the UTX stabilization domain was sufficient to restabilize UTX and recover gene expression. [Rickels 2020] Though its catalytic activity appears dispensable, MLL4 itself is essential to embryonic development, and is required for the transition from naïve to poised pluripotency in mESCs. However, we found that the effects of MLL4 loss could be rescued by depletion of the opposing H3K4 demethylase LSD1, which is a member of co-repressive complexes with additional deacetylase activity. [Cao 2018] Together, these studies suggest that supportive or antagonistic interactions with histone lysine demethylases may be a common non-catalytic function for histone methyltransferases. The noncatalytic activities of histone modifying enzymes are important to processes underlying development and disease. For example, after finding that SET1B primarily localized to the cytoplasm in human cell lines, we also found that knockout of the SET1B catalytic SET domain (SET1B DSET) could not recapitulate the loss of cell viability caused by SET1B knockdown. Instead, the stabilizing interaction with its cytoplasmic partner BOD1 appears to be the essential function of SET1B that allows for cytoplasmic COMPASS “moonlighting.” Further, the effect of SET1B knockdown can largely be rescued by additional knockdown of key proteins within the lipid biosynthesis-stimulating Adiponectin pathway, indicating that cytoplasmic COMPASS moonlighting negatively regulates lipid biosynthesis. [Wang 2017] In another study, we investigated the H3K79 methyltransferase DOT1L, which is required for mammalian development and implicated in cancer. We found that loss of its catalytic activity could not recapitulate the effect of total DOT1L loss on establishment of the neural progenitor expression signature or glial cell specification. The DOT1L complex (DotCom) shares subunits with the super elongation complex (SEC), and we showed that DOT1L similarly functions to promote productive elongation, independent of its catalytic activity.[Cao 2020] 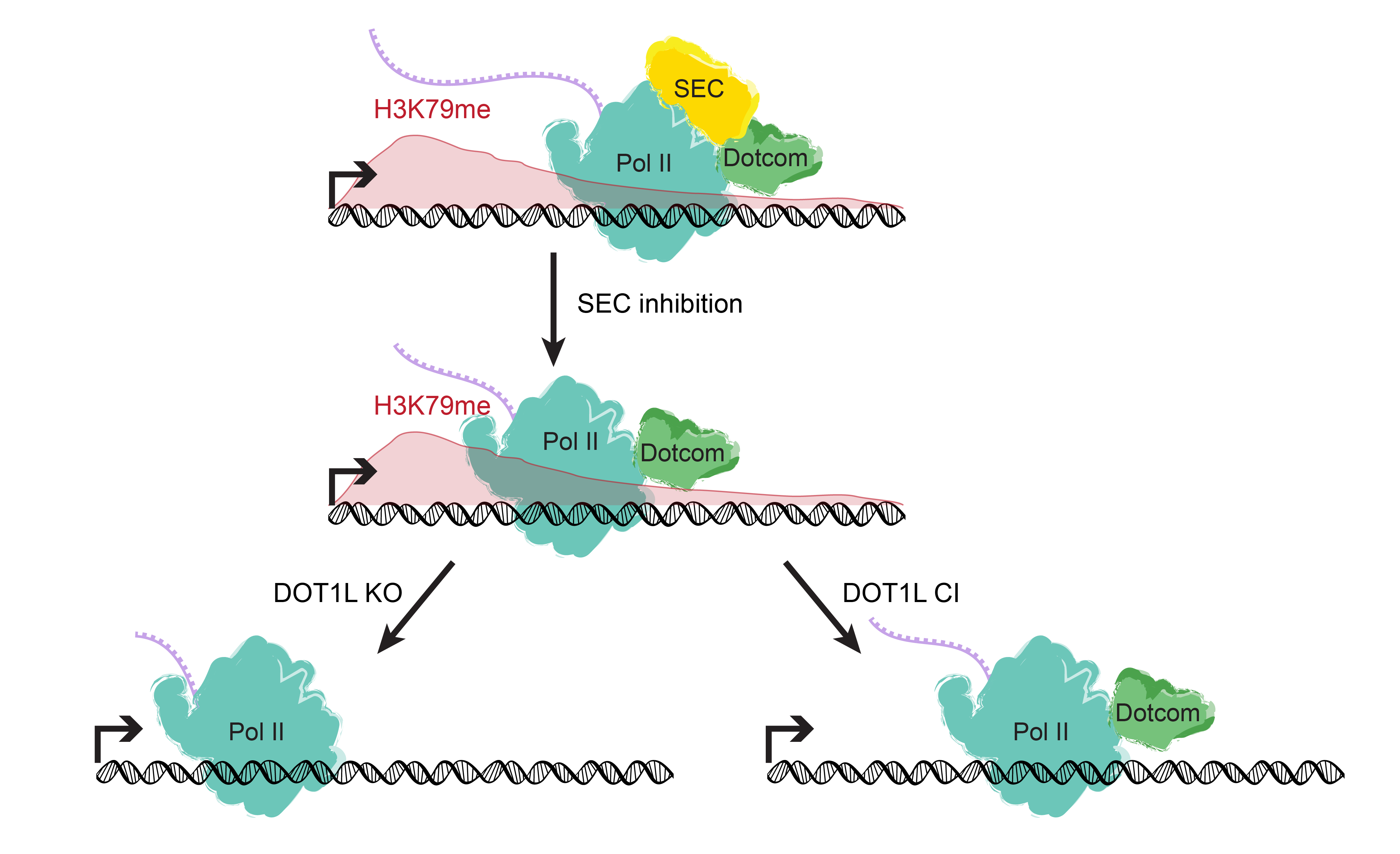
In addition to revealing non-catalytic functions for histone modifying enzymes, our studies with catalytic-dead methyltransferase mutants raise an important question: if the enzymatic activity of a histone lysine methyltransferase (or other histone modifier) is not required for cellular or even organismal viability, then why is the enzymatic activity there at all? What is its functional role? We have found some evidence of gene- or context-specific requirement for histone lysine methyltransferase activity. For example, we found that otherwise viable and self-renewing mESCs with catalytic-dead MLL2 display defects in differentiation, and we demonstrated that both the methyltransferase activity of MLL2 and the chromatin-targeting association of its own CXXC domain with CpG-rich enhancers are both required for the activation of a subset of primordial germ cell (PGC) specification genes. [Hu 2017]
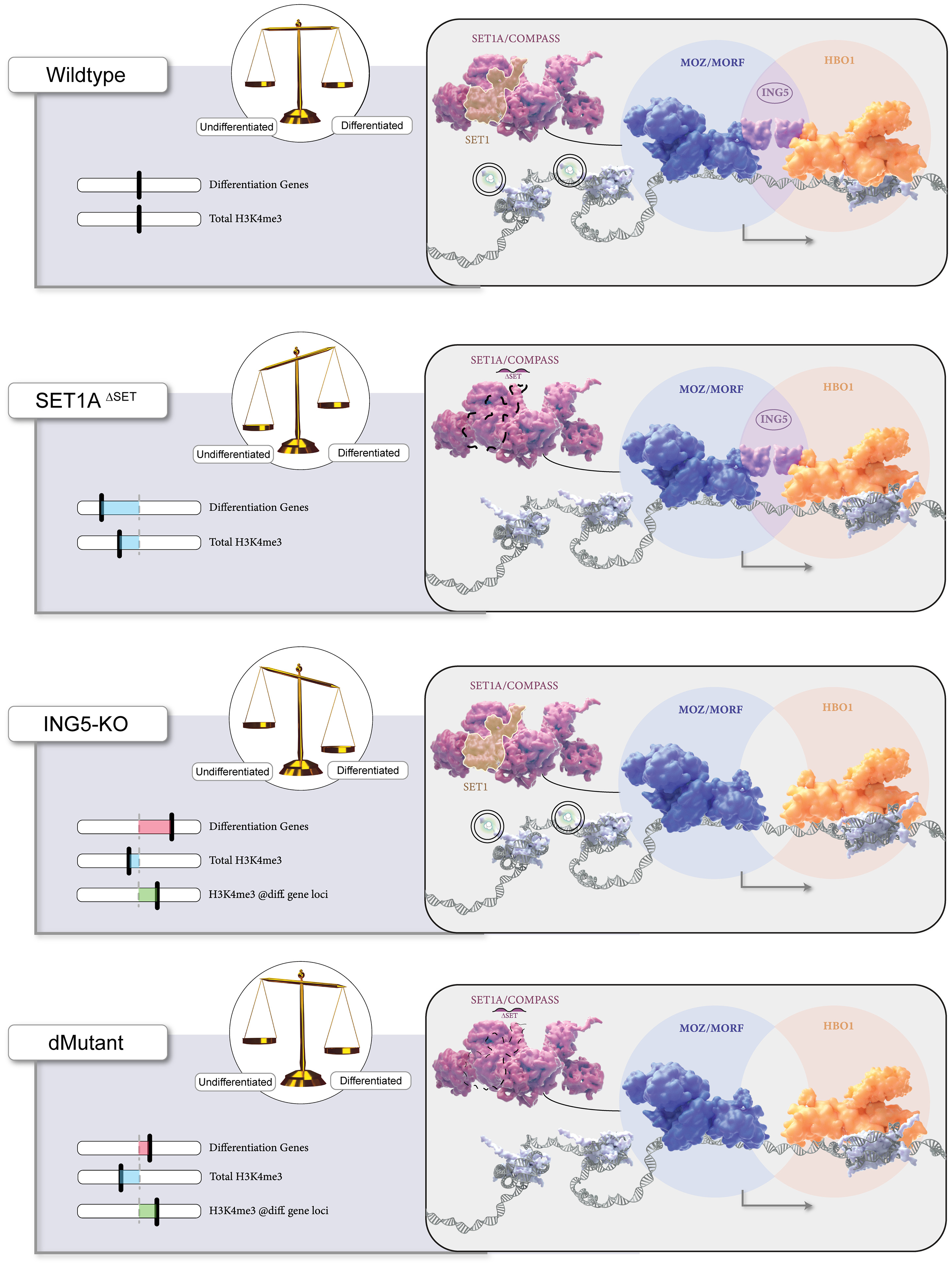
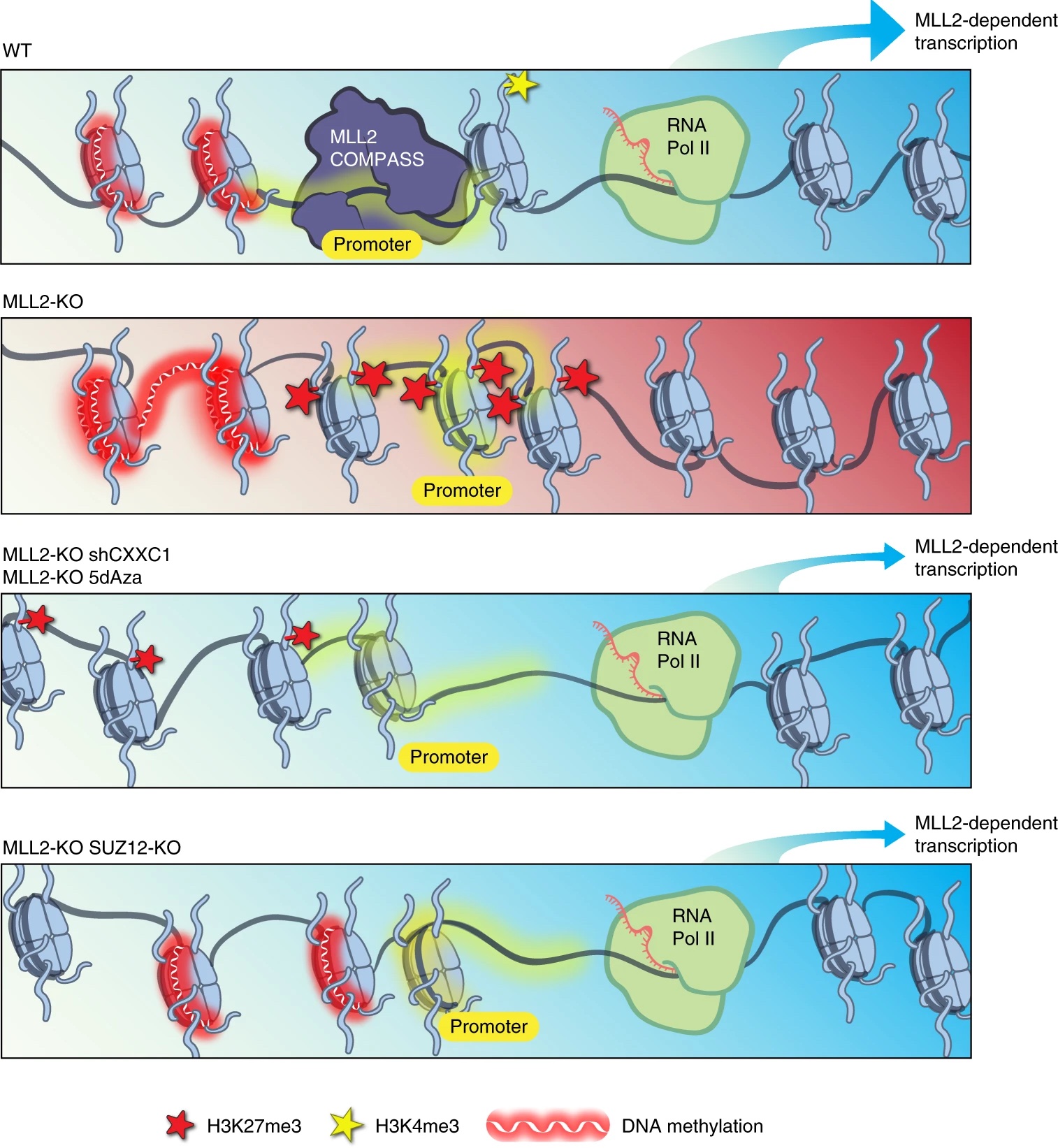
We have also found that otherwise viable and self-renewing SET1A DSET (catalytic-dead SET1A) mESCs display defects in differentiation, and we showed that methyltransferase activity is required for activating transcription from bivalent promoters of lineage-specific genes. Though MLL2 also deposits H3K4me3 at these promoters, our findings suggested that SET1A is specifically required in the context of the transition from pluripotent self-renewal to lineage-specific differentiation. [Sze 2017] In a recent follow up study, we showed that perturbation of the catalytic SET domain causes a phenotype distinct from that of Set1A null mice, with lethality at a later embryonic stage. Then, when we screened SET1ADSET mESCs for genetic dependencies of catalytic SET domain perturbation, we identified the MOZ/MORF and HBO1 acetyltransferase complex member ING5 as a potential Set1A/COMPASS partner coregulating ESC pluripotency.[Cenik 2022] Importantly, multiple histone modifying enzymes can all deposit the same mark, so that balance between these enzymes and other chromatin modifiers may underly the differential gene- and context-specific function of independent histone-modifying activities. When we examined the effects of mutating different COMPASS methyltransferases on H3K4me3 deposition, we were able to demonstrate a functional redundancy between SET1A and SET1B, which appear to be the primary depositors in mESCs, complemented by the activity of MLL2. [Sze 2020] Unlike MLL2, which has its own chromatin-targeting CXXC domain, SET1A and SET1B instead make use of a separate CXXC1 subunit to target promoter regions. Interestingly, we found that knockdown of the SET1A/B COMPASS subunit CXXC1 rescued lost gene expression in MLL2 KO mESCs, despite the failure of this knockdown to restore lost H3K4me3. We further determined that in the absence of MLL2 from its target loci, the presence of SET1A/B excluded Polycomb histone methyltransferase activity, enabling repressive DNA methylation by DNMT1. [Douillet 2020]
Watch Video
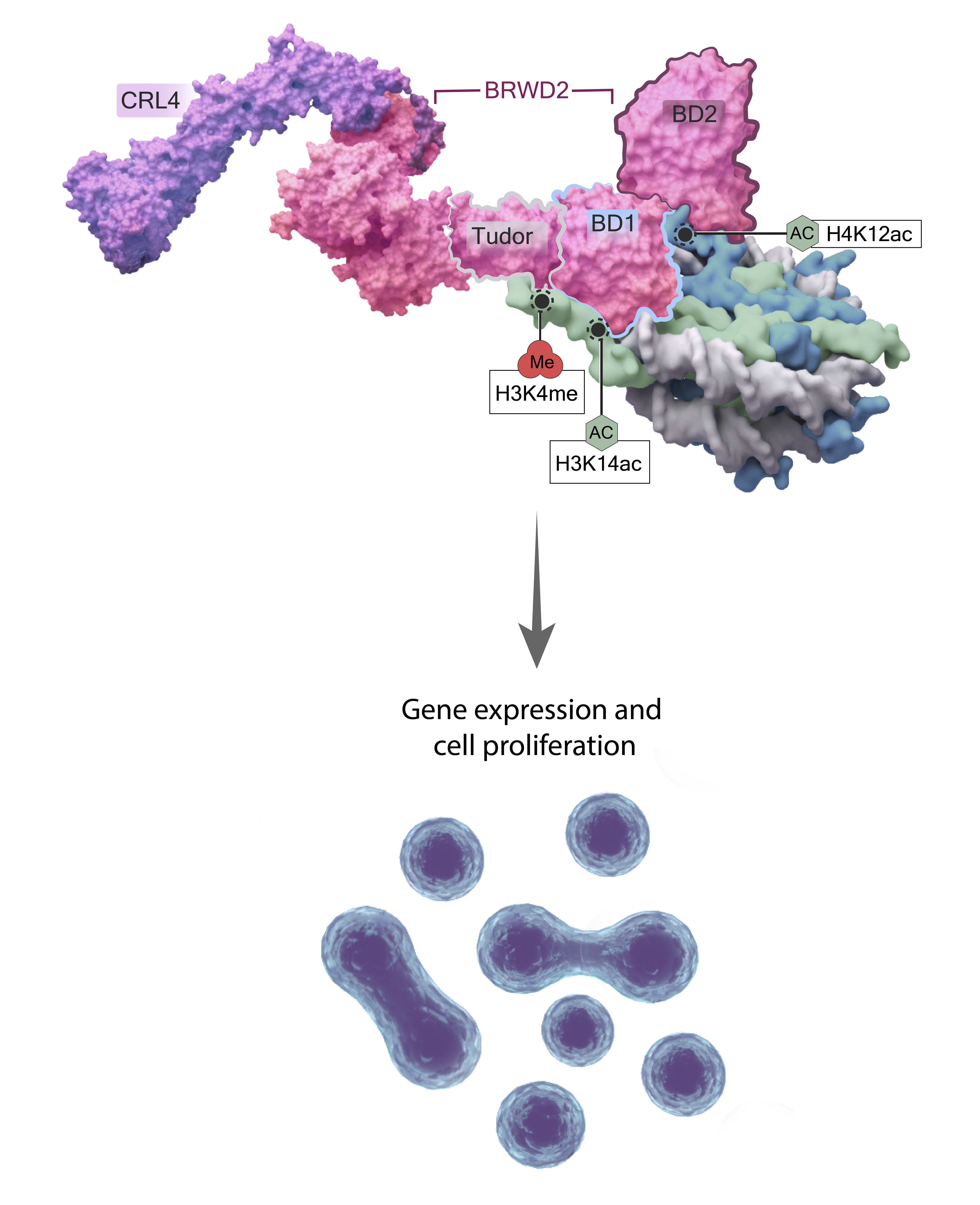
What about the molecular function of histone modifications? Histone post-translational modifications are specifically recognized and bound by chromatin-associated proteins often referred to as “readers,” and our lab has elucidated some of these often complex interactions. For example, we identified and characterized the conserved chromatin protein factor PHIP (BRWD2), which recognizes H3K4me. We found that nucleosomes, chromatin remodelers and the CRL4 complex all associate with PHIP, that it binds H3K4me via a previously unidentified crypto-Tudor domain, and that the activity of COMPASS family methyltransferases differentially regulate its chromatin occupancy. [Morgan 2017] Multivalent chromatin binding is difficult to identify in bulk ChIP-seq data, which can’t determine if two (or more) marks actually coexist on same histone tail. However, in a follow-up study using single cell-derived colonies, we were able to show that PHIP not only recognizes H3K4me, but in fact binds nucleosomes via a trivalent BD1/BD2/Tudor reader domain that recognizes H3K4meK14ac/H4K12ac as its cognate modification signal: the BD1 domain binds H3K14ac, BD2 binds H4K12ac and the Tudor domain binds H3K4me.) [Morgan 2021] You can read more about our gene- and context-specific re-evaluation of histone modifications and moonlighting roles for histone modifying enzymes in our recent Perspective in Science Advances [Morgan and Shilatifard, 2023] As always, we welcome potential students, fellows and early career scientists interested in performing rigorous biochemical and molecular studies that challenge existing paradigms. Active Areas of Investigation